Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Convolutional Neural Networks Based Paddy Leaf Disease Detection
Authors: M. Poornima, S. Bayanna, T. Manasa, Shaik Mahammad Arshu, V. Jayaprakash Reddy, T. Haribabu
DOI Link: https://doi.org/10.22214/ijraset.2024.60546
Certificate: View Certificate
Abstract
This study proposes a novel approach for detecting paddy leaf diseases through the integration of Convolutional Neural Networks (CNNs). The process begins by converting input RGB images into HSV color space, where the saturation component is extracted. Subsequently, a binary image is generated, followed by background removal to enhance the accuracy of segmentation. The background-removed image is then converted back to the HSV color space for further processing. K-means segmentation is applied to segment the leaf regions effectively. The CNN architecture is employed for classification, categorizing the detected regions into various disease classes such as Healthy, Sheath Rot, Bacterial Leaf Blight, Brown Spot, and Leaf Blast. Additionally, a voice output feature is incorporated to provide audible feedback on the detected diseases. The study focuses on evaluating the accuracy of disease classification, aiming to demonstrate the effectiveness of the CNN approach in improving performance and reducing misclassifications compared to traditional methods. Overall, the proposed methodology offers a promising solution for accurate and efficient paddy leaf disease detection, with potential applications in agricultural management and crop protection.
Introduction
I. INTRODUCTION
One of the most important staple crops in the world, rice accounts for a large amount of the global food supply. Paddy farming, however, is fraught with difficulties. One such issue is the presence of leaf-related illnesses, which can negatively influence both output and quality. For disease management in paddy fields to be efficient, early detection and prompt response are essential. Conventional disease detection techniques mostly rely on manual inspection, which is time-consuming, labor-intensive, and frequently subjective. Automated systems based on Convolutional Neural Networks (CNNs) have emerged as viable methods for plant disease detection with recent advances in computer vision and machine learning. CNNs are a subclass of deep learning models that are specifically tailored to image identification applications. They can extract intricate features and patterns from unprocessed pixel data.
Regarding the identification of paddy leaf disease, CNN-based techniques have a number of benefits over conventional techniques. Their ability to quickly evaluate vast amounts of leaf picture data makes early detection and action possible. CNNs can also offer impartial and consistent evaluations, which lessens the need for human knowledge. An extensive overview of a Convolutional Neural Network (CNN) based method for paddy leaf disease detection is presented in this research. The suggested approach uses digital photos of paddy leaves to analyze diseases and automate the procedure. Image preprocessing, feature extraction, and classification using a CNN model trained on a collection of annotated leaf pictures are some of the crucial elements in the methodology.
Plant diseases present greater challenges for the monitoring procedure when done manually. The manual method requires greater processing time, a significant amount of labor, and knowledge of plant diseases. In order to identify plant diseases, image processing techniques are applied. Experts typically use their unaided eyes to manually identify plant diseases, which takes more time and money on large farms. It is challenging to comprehend and occasionally results in a mistake when determining the type of disease. In recent years, rice production has decreased due to a lack of knowledge on appropriate management practices for treating leaf diseases in rice plants. In order to address this, a suitable and quick rice leaf disease recognition system is required. The use of automatic techniques for plant disease detection is advantageous because it minimizes the amount of work involved in monitoring large-scale agricultural farms and identifies disease symptoms early on, when they first manifest on plant leaves.
Unusual growth or indicators of the pathogen, such as bacterial slime (an outward manifestation of a disease known as bacterial wet wood) or insect larvae that emerge from eggs and feed on leaves, can be used to identify diseased plants.
In order to identify plant diseases, farmers and gardeners frequently consult literature and the internet. Additionally, they can bring a small cutting of an affected plant—or a picture of it—to a nearby garden center, where the staff can frequently assist in diagnosing and treating the illness. Cuttings of the plant may be examined by scientists in labs if a disease is difficult to diagnose. Agar plates can be used to cultivate pathogens, and controlled environments can be used to cultivate viruses. After that, cultivated bacteria and viruses are identified by biochemical assays, often with the aid of diagnostic kits that include monoclonal antibodies.
This study's organizational framework separates the research into several areas. Section 2 presents the Literature Survey. The topics of current and suggested system approaches were covered in sections three and four. Additionally, section 5 discusses the simulation results that are displayed, and section 6 concludes with a presentation of future work and conclusions. give suitable method for arithmetic calculations. The Vedic formulas requires less time than the regular formulas or method of calculations.
II. LITERATURE SURVEY
R. R. Atole,[1] In order to determine whether or not common rice leaves are diseased, D. Park conducted research on the detection of paddy illness using a convolutional neural network classifier.
Konstantinos Ferentinos [2] have investigated the identification and treatment of diseases in paddy crops. In order to demonstrate plant disease detection and diagnosis, Constantine et al. created a deep learning technique based on CNN models. A novel two-level neural network architecture was presented by Marco et al. for the classification of plant diseases.
Y. Lu, S. Yi, N. Zeng,[3] Y. Liu, and Y. Zhang have studied the paddy crop Recent advancements in convolutional neural networks (CNNs) based on deep learning have made it possible for researchers to greatly increase the classification accuracy of images. This paper describes a deep learning-based method for identifying pests and illnesses in rice using photos taken in actual environments with a variety of backgrounds.
Y. Es-saady, T. El Massi, M. El Yassa ,D. Mammass , and A. Benazoun,[4] have investigated the detection of diseases in paddy crops. This research describes a machine vision system that uses photos to automatically identify plant leaf diseases. Two of his SVM classifiers' serial combination technique serves as the foundation for the suggested system. The first classifier uses color to categorize images. At this point, illnesses with comparable symptoms or colors are thought to be in the same class. Subsequently, a second classifier is employed to differentiate between groups of colors based on their shape and texture characteristics.
III. EXISTING METHOD
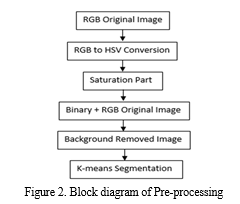
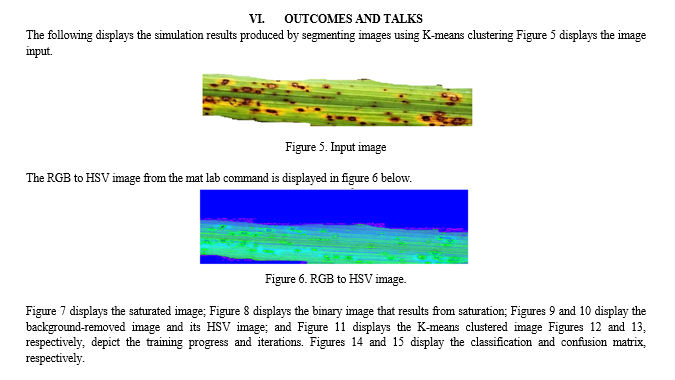
Agriculture places a high priority on the diagnosis of paddy leaf disease as it affects crop productivity and food security. For automating disease identification in paddy plants, utilizing Convolutional Neural Networks (CNNs), a subset of deep learning, in combination with sophisticated image processing methods such as RGB to HSV conversion, saturation analysis, binary image generation, background removal, and K-means segmentation, holds great potential. With the use of this creative method, illnesses may be diagnosed quickly and accurately by using CNNs' ability to recognize intricate patterns and characteristics in plant photos. Figure 1 depicts the current method's block diagram, and Figure 2 shows the pre-processing.
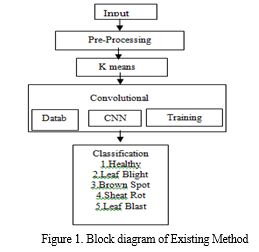
In MATLAB, K-means segmentation is a widely used method for segmenting images. Using pixel similarities, an image is divided into K clusters in this technique, where K is the desired number of segments. K cluster centroids are first chosen at random, frequently from the color space of the image. Using color similarity, each pixel is mapped to the closest centroid, forming clusters. The mean of the pixels in each cluster is then used to recalculate the centroids. Up until convergence, this centroid update and assignment procedure is repeated. As a consequence, the image is divided into segments, with each pixel assigned to a certain cluster based on similarities. K-means segmentation is used in computer vision, object recognition, and image reduction to help in effective analysis and processing of visual data.
IV. PROPOSED METHOD
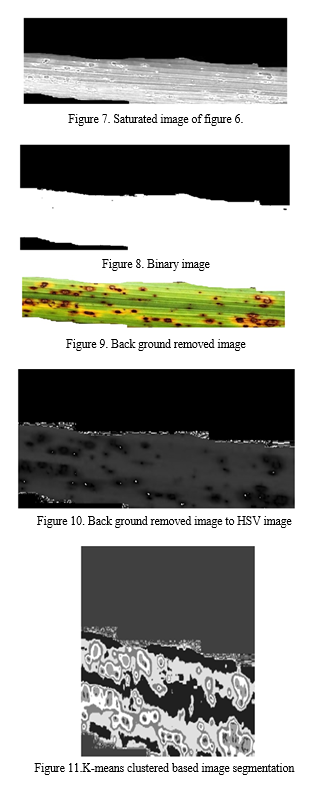
Using Convolutional Neural Networks (CNNs), we first preprocess the RGB input photos into the HSV colour space as part of our methodology for paddy leaf disease identification. Next, in order to improve the visibility of features relevant to sickness, we extract the saturation component from the HSV image. The leaf sections are then separated from the backdrop using a thresholding technique, which results in the generation of a binary image. In order to further improve the segmentation, we use techniques for background removal to get rid of undesired parts and noise.
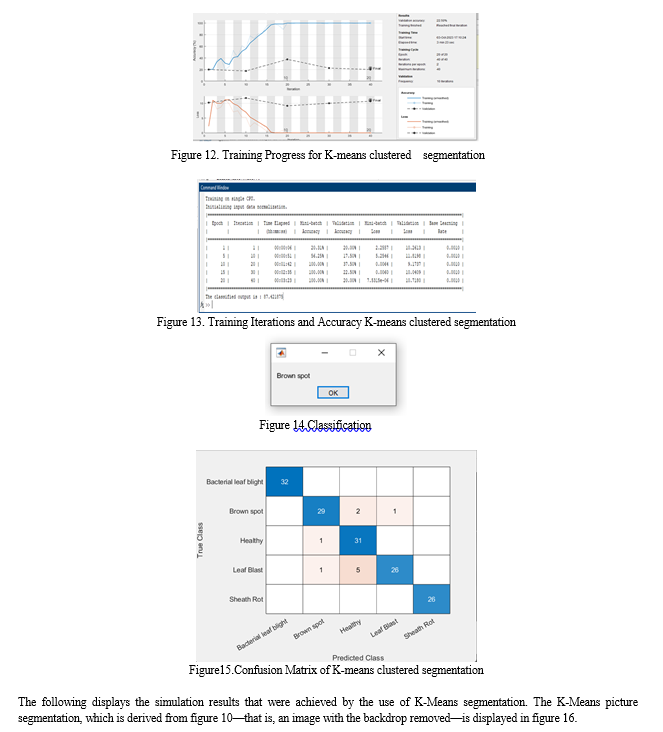
For uniformity in feature representation, the background-removed image is then transformed back to the HSV colour space. Subsequently, we employ K-means segmentation to efficiently divide the leaf sections. Our suggested CNN architecture is made up of several layers that are intended to extract hierarchical features and carry out classification, such as convolutional, pooling, and fully connected layers. Predictions for several disease classes, including Healthy, Sheath Rot, Bacterial Leaf Blight, Brown Spot, and Leaf Blast, are included in the CNN model's output. We also include a voice output feature to give an audio feedback on the illnesses identified. In order to evaluate the success of the suggested methodology, we lastly examine the accuracy of illness classification. Our goal is to show how employing a CNN strategy can improve performance and decrease misclassifications when compared to other methods. Figure 3 depicts the suggested method's block diagram, and Figure 4 shows its pre-processing.
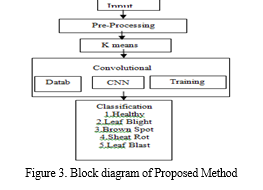
- Preprocessing: The initial handling of data to get it ready for subsequent analysis or primary processing. Any initial or preparatory processing stage where multiple processes are needed to prepare data for the user can be referred to by this phrase.
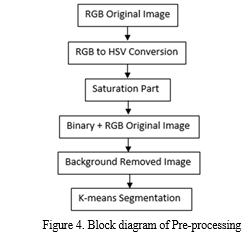
2. Procedures for using image processing to find plant diseases:
a. Users upload their own photographs to our online application or an Android smartphone creates the input images first.
b. Dividing Image enhancement, color space conversion, and segmentation are all included in pre-processing. First, a filter is used to improve the digital version of the photograph. Next, turn every picture into an array. Every image name is transformed into a binary field using the scientific term for Binarizes Diseases.
c. CNN classifiers are taught to recognize ailments specific to every group of plants. The classifier, trained to classify different plant diseases, is called up based on the Level 2 results. The leaves are categorized as "healthy" if they are absent.
- RGB2HSV Converted Image: A popular tool in MATLAB for converting an image from the RGB (Red, Green, Blue) colour space to the HSV (Hue, Saturation, Value) colour space is the `rgb2hsv` function. For many image processing jobs, particularly those involving color analysis and manipulation, this conversion is essential. While the HSV color space offers a more logical representation by separating color information (hue) from intensity (value), the RGB color space depicts colors based on the intensity of three main colors. Hue specifies the color's shade or tint, saturation determines the color's vibrancy or purity, and value indicates brightness. MATLAB's `rgb2hsv` function facilitates efficient color-based operations and analysis by accepting an RGB image as input and producing a corresponding HSV image.
- Saturation: is typically associated with the HSV (Hue, Saturation, Value) color space, where the 'Saturation' component controls the intensity of color. To convert an image to HSV color space in MATLAB, you can use the `rgb2hsv` function. Adjusting the saturation involves modifying the 'Saturation' channel while keeping the hue and value components intact. Increasing saturation intensifies colors, making them more vivid, while reducing saturation desaturates the image, turning it grayscale. This transformation is valuable in various applications, including image enhancement, artistic rendering, and color correction. MATLAB provides powerful tools for efficiently handling and manipulating saturation to achieve desired visual effects and improve image aesthetics.
- Binary Image: In MATLAB, thresholding is used to transform an image to a binary image. Based on a predetermined threshold, pixel values are divided into two groups: black (0) and white (1). First, the grayscale or color image is read using routines such as `imread () }.The `im2bw () }functions are used to assign pixel values greater than or equal to the threshold to white (1) and values below the threshold to black (0) in order to build a binary image. A threshold value is first selected.
- Image With Background Removed: A crucial preprocessing step in computer vision and image processing is background removal, which includes separating the foreground or main subject from the background in images. This process usually makes use of a variety of MATLAB techniques, including color-based segmentation, edge detection, and deep learning-based methods. For simpler backgrounds, techniques like background reduction and chroma keying can be applied.
More complex techniques make use of cutting-edge algorithms such as Grab Cut or neural networks based on U-Net, which is often used for jobs involving semantic segmentation. Accurate segmentation or masking that distinguishes the subject from the backdrop is the aim. After obtaining the mask, the original image can be blended with a transparent or background to create a clear, isolated subject that can be used for virtual reality, object detection, and image editing.
- K-Means Segmentation: In MATLAB, K-means segmentation is a widely used method for segmenting images. Using pixel similarities, an image is divided into K clusters in this technique, where K is the desired number of segments. K cluster centroids are first chosen at random, frequently from the colour space of the image. Using colour similarity, each pixel is mapped to the closest centroid, forming clusters. The mean of the pixels in each cluster is then used to recalculate the centroids. Up until convergence, this centroid update and assignment procedure is repeated. As a consequence, the image is divided into segments, with each pixel assigned to a certain cluster based on similarities. K-means segmentation is useful for computer vision, object recognition, and picture compression since it facilitates the effective analysis and processing of visual data.
Steps for implementation
First, assume that the input image is an RGB image.
Step 2: Use a MATLAB command to convert the RGB image to an HSV image.
Step 3: Extract the saturation portion of the HSV image.
Step 4: The binary picture, or black and white image, is obtained from step 3.
Step 5: Get the picture with the background erased.
Step 6: Transform the image with the background erased into an HSV image. We obtain a Hue image with watershed lines by watershed segmentation.

VII. ACCURACY DIFFERENCE
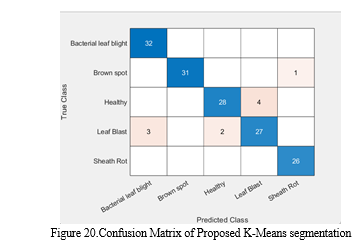
The performance accuracy for existing method is 85.4 and the performance accuracy for proposed method is 98.5.We can increase accuracy by increasing CNN layers
VII. FUTURE SCOPE
There is a great deal of potential for future research and advancement in the field of paddy leaf disease detection using Convolutional Neural Networks (CNNs). Among the possible directions for further investigation are:
Enhanced Model designs: Constantly enhancing CNN designs to get more efficiency and accuracy in the identification of diseases. To further improve performance, this may entail experimenting with novel designs, such as ensemble models, capsule networks, or attention methods.
Examining the possibility of using transfer learning methods to make use of CNN models that have already been trained on bigger datasets or similar plant species. To improve the generalization capacity of models across various geographic locations and environmental circumstances, domain adaptation techniques could also be investigated.
VIII. ACKNOWLEDGEMENT
We are really happy to present the project's preliminary report. SV College of Engineering (SVCE) (Autonomous), Tirupati, Andhra Pradesh, India: I would like to take this opportunity to thank my guide, Mrs. M. Poornima M. Tech , Assistant Professor, and Dr. D. Srinivasulu Reddy, Ph.D., Professor, & Head of the Department (HOD) of Electronics and Communication Engineering, for providing me with all the support and guidance I needed. I sincerely appreciate their thoughtful advice and support, which were quite beneficial. I'm grateful to everyone!
Conclusion
In summary, this study has shown how well Convolutional Neural Networks (CNNs) function for automating the diagnosis of diseases affecting rice leaves. With the use of a thorough workflow that consists of feature extraction, picture pre-processing, and CNN model classification, we have successfully identified a number of leaf diseases, including blight, brown spot, and sheath rot, with encouraging results. The suggested CNN-based technique performs better than conventional approaches of disease detection in terms of accuracy, speed, and scalability, according to the experimental results reported in this study. Our method uses deep learning and computer vision techniques to quickly and objectively analyze the health of paddy leaves. This allows for early diagnosis and action to minimize crop losses.
References
[1] T. Gupta, ”Plant leaf disease analysis using image processing technique with modified SVM-CS classifier,” Int. J. Eng. Manag. Technol, no. 5, pp. 11-17, 2017. [2] Y. Es-saady T. El MassiM. El Yassa, D. Mam mass, and A. Bena zoun,”Automatic recognition of plant leaves diseases based on serial combination of two SVM classifiers,” International Conference on Electrical and Information Technologies (ICEIT) pp. 561-566, 2016. [3] P. B. Padol and A. A. Yadav, “SVM classifier based grape leaf disease detection,” Conference on Advances in Signal Processing (CASP), pp. 175-179, 2016. [4] L. Liu and G. Zhou, “Extraction of the rice leaf disease image based on BP neural network,” International Conference on Computational Intelligence and Software Engineering, pp. 1-3,2009. [5] S. Arivazhagan and S.V. Ligi, “Mango Leaf Diseases Identification Using Convolutional Neural Network,” International Journal of Pure and Applied Mathematics, vol. 120,no. 6, pp. 11067-11079,2008. [6] B. Liu, Y. Zhang, D. He and Y. Li, “Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks Symmetry,” X. X. & Suen, C. Y. A novel hybrid CNN–SVM classifier for recognizing handwritten digits. Pattern Recognition vol. 45,pp. 1318–1325,2012. [7] Y. Lu, S. Yi ,N. Zeng Y. Liu, and Y. Zhang, “Identification of Rice Diseases Using Deep Convolutional Neural Networks,” Neurocomputing, 267, pp. 378-384,2017. [8] R. R. Atole, D. Park, “A Multiclass Deep Convolutional Neural Network Classifier for Detection of Common Rice Plant Anomalies,” International Journal Of Advanced Computer Science And Applications vol. 9,no. 1,pp. 67– 70,2018. [9] V. Singh, A. Misra, “Detection of Plant Leaf Diseases Using Image Segmentation and Soft Computing Techniques,” Information Processing in Agriculture, Vol. 4 ,no. 1,pp. 41–49,2017. [10] P. Konstantinos Ferentinos, “Deep Learning Models for Plant Disease Detection and Diagnosis,” Computers and Electronics in Agriculture ,vol. 145,pp. 311– 318,2018. [11] S. P. Mohanty, D. P. Hughes, M. Salath´e, “Using Deep Learning for Image-Based Plant Disease Detection,” Frontiers in plant science ,vol. 7. [12] S. Xie, T. Yang, X. Wang, and Y. Lin, “Hyper-class augmented and regularized deep learning for fine grained image classification,” IEEE Conference on Computer Vision and Pattern Recognition,CVPR2015,pp. 2645–2654,June 2015. [13] Y. Lu, S. Yi, N. Zeng, Y. Liu, Y. Zhang, ” Identification of rice diseases using deep convolutional neural networks”, Neurocomputing 267,pp. 378–384,2017. [14] K. Simonyan, A. Zisserman, “Very Deep Convolutional Networks for Large Scale Image Recognition”, Ar xiv preprint ar Xiv: arXiv,pp. 1409- 1556,2015.
Copyright
Copyright © 2024 M. Poornima, S. Bayanna, T. Manasa, Shaik Mahammad Arshu, V. Jayaprakash Reddy, T. Haribabu. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET60546
Publish Date : 2024-04-18
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

